Abstract
Microarray is one of the most recent advances being used for cancer research; provides assistance in the pharmacological approach to treat various diseases, including oral lesions. Microarray helps to analyze a large number of samples that have been previously registered or new samples; it even helps test the incidence of a particular marker in tumours. Until recently, the use of microarrays in dentistry has been very limited, but in the future, as the technology becomes affordable, its use may increase. Here, we discuss the various techniques and applications of microarrays or DNA chips.
KEYWORDS: Cancer, human genome, microarray, tissue microarray
Technique
Gene microarray technology is based on the ability to deposit many (tens of thousands) different DNA sequences on a small surface, usually a glass slide (often referred to as a “chip”). The different DNA fragments are arranged in rows and columns so that the identity of each fragment is known through its location on the array. Two types of microarrays are the gene expression microarray and the tissue microarray (TMA).
Techniques such as Northern blot and reverse transcriptase-polymerase chain reaction (RT-PCR) allow an analysis of only a few genes per experiment. But microarrays or “global expression profiling” not only analyze orders of magnitude more genes than was previously possible, but they also have the advantage that the genes examined are not influenced by gene preselection.
Microarray principle
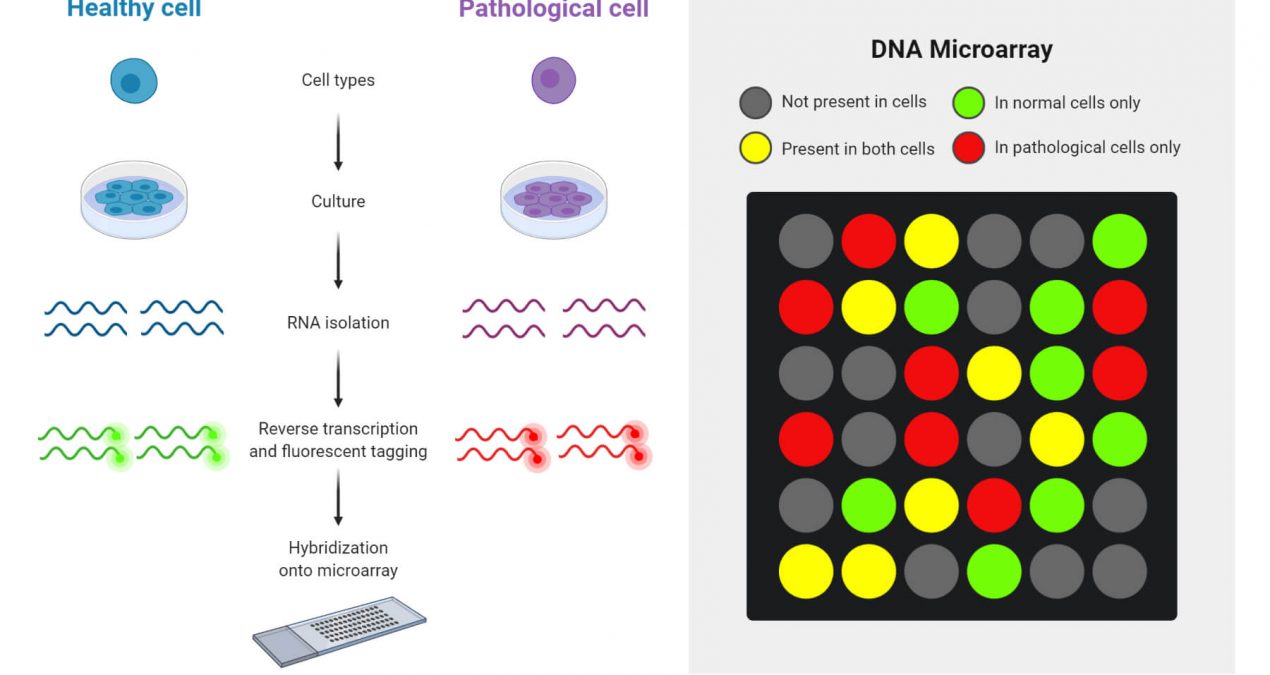
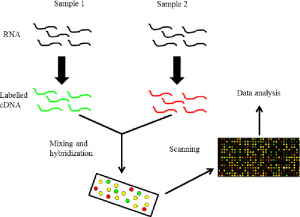
mRNA is an intermediary molecule that carries genetic information from the cell nucleus to the cytoplasm for protein synthesis. Every time some genes are expressed or in their active state, many copies of mRNA corresponding to the particular genes are produced by a process called transcription. These mRNAs synthesize the corresponding protein by translation. So indirectly by evaluating the various mRNAs, we can evaluate the genetic information or gene expression. This helps in understanding the various processes behind each altered gene expression.
Therefore, the mRNA acts as a surrogate marker. Since mRNA is easily degraded, it needs to be converted to a more stable cDNA form. The labelling of the cDNA is carried out using Cy3 (green) and Cy5 (red) fluorochrome dyes. The principle behind microarrays is that complementary sequences will bind to each other. Unknown DNA molecules are cut into fragments by restriction endonucleases; fluorescent markers are attached to these DNA fragments. They are then allowed to react with the probes on the DNA chip. The target DNA fragments along with complementary sequences then bind to the DNA probes. The remaining DNA fragments are removed by washing.
Target DNA pieces can be identified by their fluorescence emission when passing a laser beam. A computer is used to record the fluorescence emission pattern and DNA identification. This technique of using DNA chips is very fast, as well as being sensitive and specific for the identification of several DNA fragments simultaneously. The study of the expression of most, if not all, of the genes in a specimen is not based on hypotheses as most studies used to be but is called “discovery-type research” or in a way less flattering. description as “fishing expeditions”. While tumour-derived cDNA is hybridized to a chip to study gene expression levels, alterations in DNA copy number (gene amplification or deletion) can be measured by hybridizing fluorescently labelled DNA from a sample. with these chips.
TMAs are constructed by transferring paraffin-embedded tissue cores into pre-drilled holes in a recipient paraffin block. With this technique, more than 500 cores can be placed in a single block. Sections cut from TMA blocks can be used for immunohistochemistry (IHC) or in situ hybridization studies. TMAs are similar to gene expression microarrays in that they have samples arranged in rows and columns on a glass slide; they differ in that each element on the TMA slide corresponds to a single patient sample, allowing multiple patient samples to be evaluated for a single molecular marker in one experiment, whereas gene expression arrays allow the evaluation of thousands of molecular markers in a single patient sample to be tested.
Applications
- In cancer
Tumour formation involves simultaneous changes in hundreds of cells and variations in genes. The microarray can be a boon for researchers as it provides a platform for the simultaneous testing of a large set of genetic samples. It especially helps in the identification of single nucleotide polymorphisms (SNPs) and mutations, tumour classification, identification of tumour suppressor target genes, identification of cancer biomarkers, identification of genes associated with chemoresistance, and drug discovery. For example, we can compare the different patterns of gene expression levels between a group of cancer patients and a group of normal patients and identify the gene associated with that particular cancer.
Gene microarrays have been used for comparative genomic hybridization. In this technique, genomic DNA is fluorescently labelled and used to determine the presence of gene loss or amplification. Array-based comparative genomic hybridization (aCGH) has been used to map genetic abnormalities in a wide variety of tumours. , including breast carcinoma, bladder carcinoma, fallopian tube carcinoma, gastric carcinoma, melanoma, and lymphoma. Gene expression data can identify groups of cases with significantly different outcomes where routine histopathologic examination does not allow for subclassification.
Conversion of a non-invasive tumour to an invasive tumour also warrants investigation. Clark et al. studied the genetic profile of melanoma cells that became metastatic and found that one gene, RhoC, was expressed more in metastatic cells than in non-metastatic melanoma cells. Microarray-based expression profiling allows us to identify gene families as well as important molecular and cellular events that could be important in complex processes such as metastasis. Future practical applications include diagnostic and prognostic management of patients.
Physicians will be able to use microarrays during early clinical trials to confirm drug mechanisms of action and assess drug sensitivity and toxicity. They can be used to develop a new molecular taxonomy of cancer, including the grouping of cancers into prognostic groups based on gene expression profiles. Areas that can be combined with microarray technologies include disease classification or molecular phenotyping; the study of gene function in relation to gene regulatory networks or functional genomics; pharmacogenomics and developmental biology.
- Antibiotic treatment
The increase in the number of resistant bacteria and superimposed infections have led to the failure of antibiotics. The virulence of bacterial strains also affects the outcome of the disease process. In the oral cavity, where anaerobic bacteria may be the infectious agent, they are often not easy to culture, especially organisms such as actinomyces. DNA microarray analysis helps, as bacterial genomic DNA often exceeds the viability of bacteria and a diagnosis can be made using a small amount of DNA, as opposed to the large number of bacteria required for culture. In the future, an abscess sample could be sent not for culture and susceptibility testing, but for DNA microarray analysis.
- Early detection of oral precancerous lesions
Leukoplakia or whitish lesions of the oral cavity can be due to a myriad of reversible conditions. Currently, microscopic examination fails to identify the small subset of these lesions that progress to oral cancer. Identifying gene expression profiles or “genomic fingerprints” will allow clinicians to differentiate harmless white lesions from precancerous lesions or very early cancer. Recent studies have illustrated the efficacy of microarrays in oral cancer. In the future, samples taken from an incisional biopsy or brush biopsy may be sent to a laboratory for gene expression analysis. Early diagnosis and treatment of oral cancer correlate with longer survival. The identification and treatment of early premalignant and cancerous oral lesions may become one of the most valuable services to be performed in the future.
Conclusion
This review has provided a brief overview of the technique behind microarrays and the various steps involved. The technique, although currently limited in its applications due to cost, may broaden its prospects once the availability of commercial products increases. The ability to record a large number of ancient samples and analyze them for various genetic alterations helps to understand the concept of molecular biology.
Microarrays hold great promise for the analysis of diseases in the oral cavity. Oral disease classifications by DNA, RNA, or protein profiles will greatly enhance our ability to diagnose, prevent, monitor, and treat our patients. Currently, microarrays are primarily a research tool. Microarrays promise a much more enhanced, individualized, biobased standard of oral care, which will have a major clinical impact on how dentistry will be practised in the future.