DNA replication mode
A big question concerned DNA replication. The structure of the DNA double helix provided a tantalizing clue as to how copying might take place. It seemed likely that the two complementary strands of the helix might separate during replication, each serving as a template for the construction of a new matching strand. But was this really the case? Spoiler alert: The answer is yes! In this article, we’ll look at a famous experiment, sometimes called “biology’s most beautiful experiment,” which established that the basic mechanism of DNA replication is semi-conservative, that is, it produces DNA molecules that contain one new and one old strand.
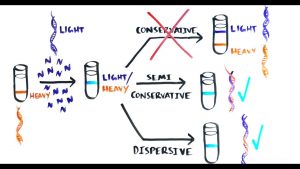
The three models for DNA replication.
There were three basic models for DNA replication that had been proposed by the scientific community after the discovery of the structure of DNA.
- Conservative. Replication produces one helix made entirely of old DNA and one helix made entirely of new DNA.
- Semiconservative. Replication produces two helices containing one old and one new DNA strand.
- Dispersive. Replication produces two helices in which the individual strands are mosaics of old and new DNA.
Most biologists at the time would probably have gone with the semi-conservative model. This model made a lot of sense given the structure of the DNA double helix, in which the two strands of DNA are perfectly and predictably complementary to each other (where one has a T, the other has an A; where one has a G, the another has a C, and so on).
This relationship facilitated the imagination of each strand by acting as a template for the synthesis of a new partner.
However, biology is also full of examples where the “obvious” solution turns out to be the wrong one. (Protein as genetic material, anyone?). Therefore, it was key to experimentally determine which model the cells actually used when replicating their DNA.
The Meselson-Stahl experiment
Matthew Meselson and Franklin Stahl were well acquainted with these three predictions and reasoned that if there was a way to distinguish old DNA from new, it should be possible to test each prediction. Aware of previous studies that had relied on isotope labels as a way to differentiate between parental and progeny molecules, the scientists decided to see if the same technique could be used to differentiate between parental and progeny DNA. If they could, Meselson and Stahl hoped they could determine which prediction and replication model was correct.
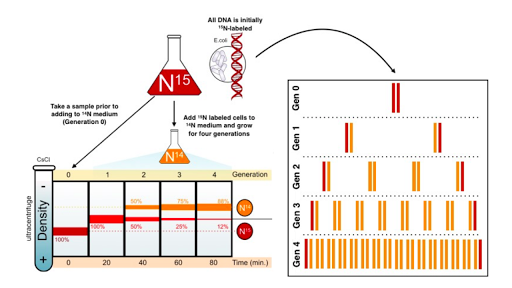
The duo, therefore, began the Meselson-Stahl experiment by choosing two isotopes of nitrogen, the common and lighter 14N, and the rarer and heavier 15N (so-called “heavy” nitrogen), as their labels and a technique known as equilibrium density. of cesium chloride (CsCl). gradient centrifugation as a sedimentation method. Meselson and Stahl chose nitrogen because it is an essential chemical component of DNA; therefore, each time a cell divides and its DNA replicates, it incorporates new N atoms into the DNA of one or both of its daughter cells, according to the correct pattern. “If several DNA species of different density are present,” they predicted, “each will form a band at the position where the density of the CsCl solution equals the buoyant density of that species. In this way, the labelled DNA with heavy nitrogen (15N) can be resolved from unlabeled DNA” (Meselson & Stahl, 1958).
The scientists then continued their experiment by growing a culture of E. coli bacteria in a medium that had the heavier 15N (in the form of 15N-labeled ammonium chloride) as its only nitrogen source. In fact, they did this for 14 bacterial generations, long enough to create a population of bacterial cells containing only the heavier isotope (by which time all of the original 14N-containing cells had died). They then changed the medium to one containing only 14N-labeled ammonium salts as the sole nitrogen source. So from then on, each new strand of DNA would be built with 14N instead of 15N.
Just prior to the addition of 14N and periodically thereafter, as the bacterial cells grew and replicated, Meselson and Stahl took DNA samples to use in equilibrium density gradient centrifugation to determine how much 15N (from the DNA original or old) versus 14N (from the new DNA) was present. For the centrifugation procedure, they mixed the DNA samples with a caesium chloride solution and then centrifuged them long enough to allow the heavier 15N and lighter 14N DNA to migrate to different positions in the centrifuge tube.
Using centrifugation, the scientists found that DNA composed entirely of 15N-labeled DNA (i.e., DNA collected just before changing the culture from one containing only 15N to one containing only 14N) formed a single distinct band because both chains were made entirely in the “heavy” nitrogen medium. After a single round of replication, the DNA reverted to a single distinct band, but the band was located at a different position along the spin gradient. Specifically, it was found midway between where all 15N DNA and all 14N DNA would have migrated; in other words, halfway between “heavy” and “light”. Based on these findings, the scientists were able to immediately exclude the conservative model of replication as a possibility.
After all, if DNA were to replicate conservatively, there should be two distinct bands after a single round of replication; half of the new DNA would have migrated to the same position as before the culture was transferred to medium containing 14N (i.e. to the “heavy” position), and only the other half would have migrated to the new position (i.e., to the “light” position). That left scientists with only two options: either the DNA replicated semi conservatively, as Watson and Crick had predicted, or it replicated dispersively.
To tell the difference between the two, Meselson and Stahl had to let the cells divide again and then sample the DNA after the second round of replication. After that second round of replication, the scientists found that the DNA separated into two distinct bands: one at a position where DNA containing only 14N would be expected to migrate, and the other at a position where DNA containing only 14N would be expected to migrate. hybrid (containing half 14N and a half 15N)) would be expected to migrate.
The scientists continued to see the same two bands after several subsequent rounds of replication. These results were consistent with the semiconservative model of replication and the reality that, when DNA replicated, each new double helix was made up of one old and one new strand. If the dispersive model were the correct model, then scientists would still have observed only one band after each round of replication.